A Platform for Exploring Subcellular Spatial Phenotypes

Sophia Mädler
Researcher, Max Planck Institute of Biochemistry (MPIB), Proteomics and Signal Transduction – Matthias Mann Lab
Read BioAfter completing her Bachelor's degree in Molecular Biomedicine at the University of Bonn, Sophia relocated to Munich to pursue her Master's studies. During this time, she gained experience in cell biology, immunology, and computational biology during internships with Stefan Canzar and at Hoffmann La-Roche in Basel. Sophia subsequently started her doctorate studies in the lab of Matthias Mann at the Max Planck Institute of Biochemistry. Here, her focus shifted to developing AI-powered tools to understand phenotypic variation in single-cell images. She developed SPARCS, a pipeline for genome-scale genetic screens on information-rich image-based phenotypes. Presently, Sophia is actively seeking job opportunities after completing her PhD.
CloseExploring subcellular spatial phenotypes with SPARCS
A platform for genome-scale microscopy-based genetic screens
See the power of spatially resolved CRISPR screening (SPARCS), which uses automated high-speed laser microdissection to reveal interesting subcellular phenotypes at the human genome scale.
In this webinar, you will learn:
- How SPARCS enables isolation of phenotypic variants in situ from virtually unlimited library sizes
- Deep learning methods to identify new phenotypes from single-cell images
- What the integration of powerful microscopy methods and CRISPR promises for genome-wide forward genetic screening
Background
Genetics seeks to understand how genes determine an organism’s phenotype. Forward genetic screening has revolutionized this pursuit by generating unbiased genotype–phenotype maps. Through the creation of genetic mutant libraries and the subsequent isolation of interesting phenotypic variants for genotyping, researchers discover connections between genes and their resulting traits in an unbiased manner. Accelerated by techniques for targeted gene disruption, this methodology now scales to genome-wide screening. So far, these screens have been limited to investigating simple phenotypes that inherently cause clonal enrichment or result in changes in fluorescence intensity measured by FACS.
Increasingly powerful microscopic imaging provides information-rich data on diverse cellular phenotypes. When coupled with recent advances in deep learning, this presents an ideal technology to read out biological phenotypes of interest in genetic screens. However, due to technological constraints, it has, up to now, found limited application.
About this webinar
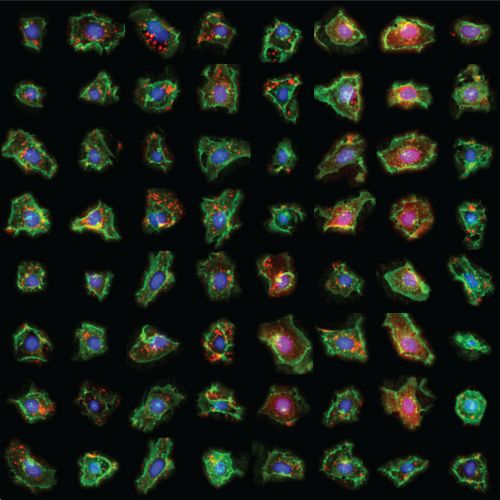
In this webinar, Sophia Mädler will present spatially resolved CRISPR screening (SPARCS), a platform for microscopy-based genetic screening for spatial subcellular phenotypes at the human genome scale. SPARCS uses automated high-speed laser microdissection to isolate phenotypic variants in situ from virtually unlimited library sizes. Using deep learning, phenotypically interesting cells can be identified based on single-cell images. To demonstrate the potential of SPARCS, Sophia will present the results from a genome-wide CRISPR-KO screen on autophagosome formation in 40 million human cells.
Phenotypic variants showing defects in autophagosome formation were identified using a convolutional neural network (CNN) classifier, which recognizes differences in the subcellular distribution of the autophagosome marker LC3B based on single-cell images. Using this classifier, SPARCS recovered almost all known macroautophagy genes in a single experiment and discovered a role for the ER-resident protein EI24 in autophagosome biogenesis. With its focus on open and accessible design, SPARCS can integrate seamlessly with any microscope. Coupled with newly developed open-source Python libraries, SPARCS allows for the rapid generation and analysis of single-cell images. Using state-of-the-art deep learning methods, cells can be characterized based on their subcellular spatial phenotypes. Identified cells of interest can then be rapidly translated into a cutting map for laser microdissection.
Throughout the webinar, Sophia will outline how SPARCS and its developed tools can be used to harness the full power of advanced imaging technologies to enable genome-wide forward genetic screening for diverse spatial subcellular phenotypes in situ.
Further Reading
More information about the SPARCS platform can be found in the following pre-print publication: SPARCS, a platform for genome-scale CRISPR screening for spatial cellular phenotypes (2023)